Dna Structure And Chemistry 392g
This document was ed by and they confirmed that they have the permission to share it. If you are author or own the copyright of this book, please report to us by using this report form. Report 3b7i
Overview 3e4r5l
& View Dna Structure And Chemistry as PDF for free.
More details w3441
- Words: 1,346
- Pages: 37
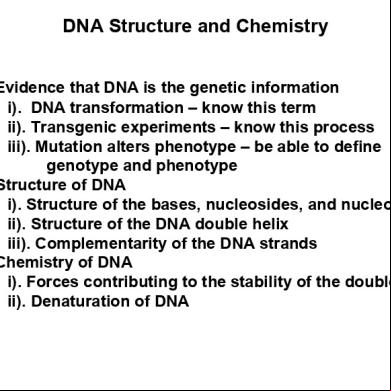
DNA Structure and Chemistry a). Evidence that DNA is the genetic information i). DNA transformation – know this term ii). Transgenic experiments – know this process iii). Mutation alters phenotype – be able to define genotype and phenotype b). Structure of DNA i). Structure of the bases, nucleosides, and nucleotides ii). Structure of the DNA double helix iii). Complementarity of the DNA strands c). Chemistry of DNA i). Forces contributing to the stability of the double helix ii). Denaturation of DNA
THE FLOW OF GENETIC INFORMATION
DNA
2
RNA
3
PROTEIN
1 DNA 1. REPLICATION (DNA SYNTHESIS) 2. TRANSCRIPTION (RNA SYNTHESIS) 3. TRANSLATION (PROTEIN SYNTHESIS)
Structures of the bases Purines
Pyrimidines
Adenine (A)
Thymine (T)
5-Methylcytosine (5mC)
Guanine (G)
Cytosine (C)
Nucleoside
[structure of deoxyadenosine]
Nucleotide
Nomenclature
Base
Nucleoside +deoxyribose
Purines adenine guanine
adenosine guanosine
hypoxanthine
inosine
Pyrimidines thymine cytosine
thymidine cytidine +ribose
uracil
Nucleotide +phosphate
uridine
ii). Structure the Structure of theofDNA DNA doublechain helix polynucleotide
5’
3’ • polynucleotide chain • 3’,5’-phosphodiester bond
A-T base pair Hydrogen bonding of the bases
G-C base pair Chargaff’s rule: The content of A equals the content of T, and the content of G equals the content of C in double-stranded DNA from any species
Double-stranded DNA 5’
3’
Major groove Minor groove
“B” DNA 3’
5’
3’
5’
Chemistry of DNA Forces affecting the stability of the DNA double helix • hydrophobic interactions - stabilize - hydrophobic inside and hydrophilic outside • stacking interactions - stabilize - relatively weak but additive van der Waals forces • hydrogen bonding - stabilize - relatively weak but additive and facilitates stacking • electrostatic interactions - destabilize - contributed primarily by the (negative) phosphates - affect intrastrand and interstrand interactions - repulsion can be neutralized with positive charges (e.g., positively charged Na+ ions or proteins)
Charge repulsion
Stacking interactions
Charge repulsion
Model of double-stranded DNA showing three base pairs
Denaturation of DNA Double-stranded DNA
Strand separation and formation of single-stranded random coils
Extremes in pH or A-T rich regions high temperature denature first
Cooperative unwinding of the DNA strands
Electron micrograph of partially melted DNA
Double-stranded, G-C rich DNA has not yet melted
A-T rich region of DNA has melted into a single-stranded bubble
• A-T rich regions melt first, followed by G-C rich regions
Hyperchromicity
Absorbance
Absorbance maximum for single-stranded DNA Absorbance maximum for double-stranded DNA
220
260
300
The absorbance at 260 nm of a DNA solution increases when the double helix is melted into single strands.
Percent hyperchromicity
DNA melting curve 100
50
0 50
70
90
Temperature oC
• Tm is the temperature at the midpoint of the transition
Percent hyperchromicity
Tm is dependent on the G-C content of the DNA
E. coli DNA is 50% G-C
50
60
70
80
Temperature oC
Average base composition (G-C content) can be determined from the melting temperature of DNA
Genomic DNA, Genes, Chromatin a). Complexity of chromosomal DNA i). DNA reassociation ii). Repetitive DNA and Alu sequences iii). Genome size and complexity of genomic DNA b). Gene structure i). Introns and exons ii). Properties of the human genome iii). Mutations caused by Alu sequences c). Chromosome structure - packaging of genomic DNA i). Nucleosomes ii). Histones iii). Nucleofilament structure iv). Telomeres, aging, and cancer
DNA reassociation (renaturation) Double-stranded DNA
Denatured, single-stranded DNA
k2 Slower, rate-limiting, second-order process of finding complementary sequences to nucleate base-pairing
Faster, zippering reaction to form long molecules of doublestranded DNA
DNA reassociation kinetics for human genomic DNA
% DNA reassociated
Cot1/2 = 1 / k2
k2 = second-order rate constant Co = DNA concentration (initial) t1/2 = time for half reaction of each component or fraction
0
50
Kinetic fractions:
fast (repeated) intermediate (repeated)
Cot1/2 Cot1/2
slow (single-copy) Cot1/2
100
I
I
I
I I I log Cot
fast intermediate slow
I
I
I
106 copies per genome of a “low complexity” sequence of e.g. 300 base pairs
high k2
1 copy per genome of a “high complexity” sequence of e.g. 300 x 106 base pairs
low k2
Type of DNA
% of Genome
Features
Single-copy (unique)
~75%
Includes most genes 1
Repetitive Interspersed
~15%
Interspersed throughout genome between and within genes; includes Alu sequences 2 and VNTRs or mini (micro) satellites Highly repeated, low complexity sequences usually located in centromeres and telomeres
Satellite (tandem) 0
~10%
fast ~10% Alu sequences are about 300 bp in length and are repeated about 300,000 times in the genome. They can be found adjacent to or within genes in introns or nontranslated regions. 2
intermediate ~15% 50 slow (single-copy) ~75% 100 1
I
I
I
I
I
I
I
I
I
Some genes are repeated a few times to thousands-fold and thus would be in the repetitive DNA fraction
Classes of repetitive DNA
Interspersed (dispersed) repeats (e.g., Alu sequences) GCTGAGG
GCTGAGG
GCTGAGG
Tandem repeats (e.g., microsatellites) TTAGGGTTAGGGTTAGGGTTAGGG
Genome sizes in nucleotide pairs (base-pairs) plasmids viruses bacteria fungi plants algae insects mollusks bony fish
The size of the human genome is ~ 3 X 109 bp; almost all of its complexity is in single-copy DNA.
amphibians reptiles
The human genome is thought to contain ~30,000 to 40,000 genes. 104
105
106
107
birds mammals 108
109
1010
1011
Gene structure promoter region
exons (filled and unfilled boxed regions)
+1 introns (between exons) transcribed region mRNA structure 5’
3’ translated region
The (exon-intron-exon)n structure of various genes histone total = 400 bp; exon = 400 bp β -globin total = 1,660 bp; exons = 990 bp HGPRT (HPRT)
total = 42,830 bp; exons = 1263 bp
factor VIII total = ~186,000 bp; exons = ~9,000 bp
Properties of the human genome Nuclear genome • the haploid human genome has ~3 X 109 bp of DNA • single-copy DNA comprises ~75% of the human genome • the human genome contains ~30,000 to 40,000 genes • most genes are single-copy in the haploid genome • genes are composed of from 1 to >75 exons • genes vary in length from <100 to >2,300,000 bp • Alu sequences are present throughout the genome Mitochondrial genome • circular genome of ~17,000 bp • contains <40 genes
Alu sequences can be “mutagenic” Familial hypercholesterolemia • autosomal dominant • LDL receptor deficiency
From Nussbaum, R.L. et al. "Thompson & Thompson Genetics in Medicine," 6th edition (Revised Reprint), Saunders, 2004.
LDL receptor gene Alu repeats present within introns
4
5 6 Alu repeats in exons
unequal crossing over
4
Alu
5
Alu
6
X 4
Alu
5
Alu
6 one product has a deleted exon 5
4
Alu
(the other product is not shown)
6
Chromatin structure
EM of chromatin shows presence of nucleosomes as “beads on a string”
Nucleosome structure
Nucleosome core (left) • 146 bp DNA; 1 3/4 turns of DNA • DNA is negatively supercoiled • two each: H2A, H2B, H3, H4 (histone octomer) Nucleosome (right) • ~200 bp DNA; 2 turns of DNA plus spacer • also includes H1 histone
Histones (H1, H2A, H2B, H3, H4) • small proteins • arginine or lysine rich: positively charged • interact with negatively charged DNA • can be extensively modified - modifications in general make them less positively charged Phosphorylation Poly(ADP) ribosylation Methylation Acetylation Hypoacetylation by histone deacetylase (facilitated by Rb) “tight” nucleosomes assoc with transcriptional repression Hyperacetylation by histone acetylase (facilitated by TFs) “loose” nucleosomes assoc with transcriptional activation
Nucleofilament structure
Condensation and decondensation of a chromosome in the cell cycle
Telomeres are protective “caps” on chromosome ends consisting of short 5-8 bp tandemly repeated GC-rich DNA sequences, that prevent chromosomes from fusing and causing karyotypic rearrangements.
Telomeres and aging Metaphase chromosome
telomere
centromer e
telomere structure
telomere <1 to >12 kb (TTAGGG)many
(TTAGGG)few
young
senescent
• telomerase (an enzyme) is required to maintain telomere length in germline cells • most differentiated somatic cells have decreased levels of telomerase and therefore their chromosomes shorten with each cell division
Class Assignment (for discussion on Sept 9th ) Botchkina GI, et al. “Noninvasive detection of prostate cancer by quantitative analysis of telomerase activity.” Clin Cancer Res. May 1;11(9):3243-3249, 2005 PDF of article is accessible on the website
THE FLOW OF GENETIC INFORMATION
DNA
2
RNA
3
PROTEIN
1 DNA 1. REPLICATION (DNA SYNTHESIS) 2. TRANSCRIPTION (RNA SYNTHESIS) 3. TRANSLATION (PROTEIN SYNTHESIS)
Structures of the bases Purines
Pyrimidines
Adenine (A)
Thymine (T)
5-Methylcytosine (5mC)
Guanine (G)
Cytosine (C)
Nucleoside
[structure of deoxyadenosine]
Nucleotide
Nomenclature
Base
Nucleoside +deoxyribose
Purines adenine guanine
adenosine guanosine
hypoxanthine
inosine
Pyrimidines thymine cytosine
thymidine cytidine +ribose
uracil
Nucleotide +phosphate
uridine
ii). Structure the Structure of theofDNA DNA doublechain helix polynucleotide
5’
3’ • polynucleotide chain • 3’,5’-phosphodiester bond
A-T base pair Hydrogen bonding of the bases
G-C base pair Chargaff’s rule: The content of A equals the content of T, and the content of G equals the content of C in double-stranded DNA from any species
Double-stranded DNA 5’
3’
Major groove Minor groove
“B” DNA 3’
5’
3’
5’
Chemistry of DNA Forces affecting the stability of the DNA double helix • hydrophobic interactions - stabilize - hydrophobic inside and hydrophilic outside • stacking interactions - stabilize - relatively weak but additive van der Waals forces • hydrogen bonding - stabilize - relatively weak but additive and facilitates stacking • electrostatic interactions - destabilize - contributed primarily by the (negative) phosphates - affect intrastrand and interstrand interactions - repulsion can be neutralized with positive charges (e.g., positively charged Na+ ions or proteins)
Charge repulsion
Stacking interactions
Charge repulsion
Model of double-stranded DNA showing three base pairs
Denaturation of DNA Double-stranded DNA
Strand separation and formation of single-stranded random coils
Extremes in pH or A-T rich regions high temperature denature first
Cooperative unwinding of the DNA strands
Electron micrograph of partially melted DNA
Double-stranded, G-C rich DNA has not yet melted
A-T rich region of DNA has melted into a single-stranded bubble
• A-T rich regions melt first, followed by G-C rich regions
Hyperchromicity
Absorbance
Absorbance maximum for single-stranded DNA Absorbance maximum for double-stranded DNA
220
260
300
The absorbance at 260 nm of a DNA solution increases when the double helix is melted into single strands.
Percent hyperchromicity
DNA melting curve 100
50
0 50
70
90
Temperature oC
• Tm is the temperature at the midpoint of the transition
Percent hyperchromicity
Tm is dependent on the G-C content of the DNA
E. coli DNA is 50% G-C
50
60
70
80
Temperature oC
Average base composition (G-C content) can be determined from the melting temperature of DNA
Genomic DNA, Genes, Chromatin a). Complexity of chromosomal DNA i). DNA reassociation ii). Repetitive DNA and Alu sequences iii). Genome size and complexity of genomic DNA b). Gene structure i). Introns and exons ii). Properties of the human genome iii). Mutations caused by Alu sequences c). Chromosome structure - packaging of genomic DNA i). Nucleosomes ii). Histones iii). Nucleofilament structure iv). Telomeres, aging, and cancer
DNA reassociation (renaturation) Double-stranded DNA
Denatured, single-stranded DNA
k2 Slower, rate-limiting, second-order process of finding complementary sequences to nucleate base-pairing
Faster, zippering reaction to form long molecules of doublestranded DNA
DNA reassociation kinetics for human genomic DNA
% DNA reassociated
Cot1/2 = 1 / k2
k2 = second-order rate constant Co = DNA concentration (initial) t1/2 = time for half reaction of each component or fraction
0
50
Kinetic fractions:
fast (repeated) intermediate (repeated)
Cot1/2 Cot1/2
slow (single-copy) Cot1/2
100
I
I
I
I I I log Cot
fast intermediate slow
I
I
I
106 copies per genome of a “low complexity” sequence of e.g. 300 base pairs
high k2
1 copy per genome of a “high complexity” sequence of e.g. 300 x 106 base pairs
low k2
Type of DNA
% of Genome
Features
Single-copy (unique)
~75%
Includes most genes 1
Repetitive Interspersed
~15%
Interspersed throughout genome between and within genes; includes Alu sequences 2 and VNTRs or mini (micro) satellites Highly repeated, low complexity sequences usually located in centromeres and telomeres
Satellite (tandem) 0
~10%
fast ~10% Alu sequences are about 300 bp in length and are repeated about 300,000 times in the genome. They can be found adjacent to or within genes in introns or nontranslated regions. 2
intermediate ~15% 50 slow (single-copy) ~75% 100 1
I
I
I
I
I
I
I
I
I
Some genes are repeated a few times to thousands-fold and thus would be in the repetitive DNA fraction
Classes of repetitive DNA
Interspersed (dispersed) repeats (e.g., Alu sequences) GCTGAGG
GCTGAGG
GCTGAGG
Tandem repeats (e.g., microsatellites) TTAGGGTTAGGGTTAGGGTTAGGG
Genome sizes in nucleotide pairs (base-pairs) plasmids viruses bacteria fungi plants algae insects mollusks bony fish
The size of the human genome is ~ 3 X 109 bp; almost all of its complexity is in single-copy DNA.
amphibians reptiles
The human genome is thought to contain ~30,000 to 40,000 genes. 104
105
106
107
birds mammals 108
109
1010
1011
Gene structure promoter region
exons (filled and unfilled boxed regions)
+1 introns (between exons) transcribed region mRNA structure 5’
3’ translated region
The (exon-intron-exon)n structure of various genes histone total = 400 bp; exon = 400 bp β -globin total = 1,660 bp; exons = 990 bp HGPRT (HPRT)
total = 42,830 bp; exons = 1263 bp
factor VIII total = ~186,000 bp; exons = ~9,000 bp
Properties of the human genome Nuclear genome • the haploid human genome has ~3 X 109 bp of DNA • single-copy DNA comprises ~75% of the human genome • the human genome contains ~30,000 to 40,000 genes • most genes are single-copy in the haploid genome • genes are composed of from 1 to >75 exons • genes vary in length from <100 to >2,300,000 bp • Alu sequences are present throughout the genome Mitochondrial genome • circular genome of ~17,000 bp • contains <40 genes
Alu sequences can be “mutagenic” Familial hypercholesterolemia • autosomal dominant • LDL receptor deficiency
From Nussbaum, R.L. et al. "Thompson & Thompson Genetics in Medicine," 6th edition (Revised Reprint), Saunders, 2004.
LDL receptor gene Alu repeats present within introns
4
5 6 Alu repeats in exons
unequal crossing over
4
Alu
5
Alu
6
X 4
Alu
5
Alu
6 one product has a deleted exon 5
4
Alu
(the other product is not shown)
6
Chromatin structure
EM of chromatin shows presence of nucleosomes as “beads on a string”
Nucleosome structure
Nucleosome core (left) • 146 bp DNA; 1 3/4 turns of DNA • DNA is negatively supercoiled • two each: H2A, H2B, H3, H4 (histone octomer) Nucleosome (right) • ~200 bp DNA; 2 turns of DNA plus spacer • also includes H1 histone
Histones (H1, H2A, H2B, H3, H4) • small proteins • arginine or lysine rich: positively charged • interact with negatively charged DNA • can be extensively modified - modifications in general make them less positively charged Phosphorylation Poly(ADP) ribosylation Methylation Acetylation Hypoacetylation by histone deacetylase (facilitated by Rb) “tight” nucleosomes assoc with transcriptional repression Hyperacetylation by histone acetylase (facilitated by TFs) “loose” nucleosomes assoc with transcriptional activation
Nucleofilament structure
Condensation and decondensation of a chromosome in the cell cycle
Telomeres are protective “caps” on chromosome ends consisting of short 5-8 bp tandemly repeated GC-rich DNA sequences, that prevent chromosomes from fusing and causing karyotypic rearrangements.
Telomeres and aging Metaphase chromosome
telomere
centromer e
telomere structure
telomere <1 to >12 kb (TTAGGG)many
(TTAGGG)few
young
senescent
• telomerase (an enzyme) is required to maintain telomere length in germline cells • most differentiated somatic cells have decreased levels of telomerase and therefore their chromosomes shorten with each cell division
Class Assignment (for discussion on Sept 9th ) Botchkina GI, et al. “Noninvasive detection of prostate cancer by quantitative analysis of telomerase activity.” Clin Cancer Res. May 1;11(9):3243-3249, 2005 PDF of article is accessible on the website